Table of Contents
Asymmetric membrane builder
Created by Dr. Min-Kang Hsieh
Contact email: mkhsieh1027@gmail.com
Update (08.24.2022):
The package (v4) was constructed based on bacterial inner membrane models with phospholipids and the system with other lipids should use v5.
The packages may not be able to apply to system with glycolipids.
The system with proteins should use a limited version asym_memb_builder_v4_prot .
Please provide comments and feedbacks to improve this package. Thanks.
In step5.2, one should be able to create input for several MD simulation packages by Force Field Converter at CHARMM-GUI page (https://charmm-gui.org/?doc=input/converter.ffconverter)
where takes the step5_assembly.psf and step5_assembly.crd file generated from step5.1 with step5_assembly.str information.
The asym_memb_builder is used to generate an asymmetric lipid bilayer from two symmetric bilayers by manipulating the smaller monolayer to match the size of the larger monolayer if the size if very different (more than 5%). The package is adapted from the scripts of CHRAMM Membrane Builder, having 5 sequential steps to build a simulation box of asymmetric membrane with explicit water and ions. Briefly, after checking the input symmetric bilayer information (Step1.0) and isolating lipid, water, and ion of each leaflet (Step1.1), the script provides the potential enlarged monolayers (Step1.2) for user’s selection and recreate a half simulation box with lateral waters and ions from original molecular structure in the input symmetric membrane system (Step1.3). The asymmetric bilayer is generated at Step2 using the original input molecular structures. The simulation box size is checked at Step3 and the concentration of KCl could be modified by editing step3_size.str if user would like to update it. The asymmetric bilayer will be rechecked and isolated by individual segment (lipid, water, and ion) at Step4.1; the water box is directly adapted from the original input without change (inactive the Step4.2); and the number of ions for neutralizing system or matching the updated KCl concentration is processed at Step4.3. Then, all the updated segments (lipid, water, and ion) are assembled at Step5.1, and the input files of the asymmetric bilayer is prepared for NAMD at Step5.2. As usual CHARMM-GUI practice, the simulation is followed by step6 equilibrium and step7 production runs.

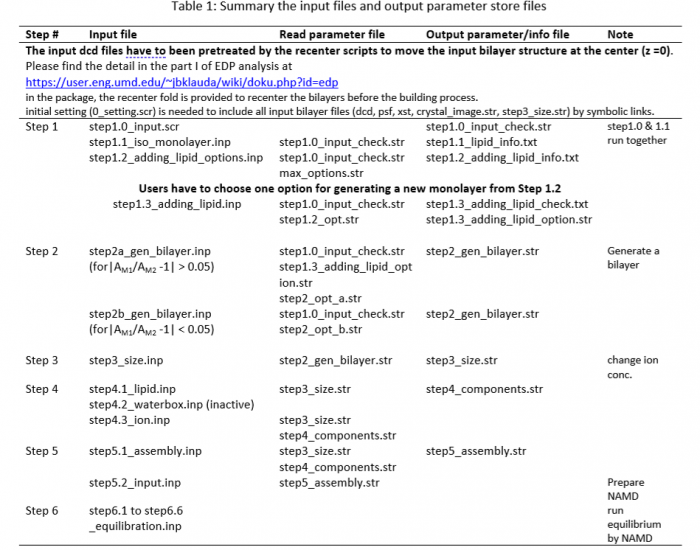
Package and operation
For DT2, the package at /lustre/mhsieh/asym_memb/asym_memb_builder_v4
/lustre/mhsieh/asym_memb/asym_memb_builder_v5.beta
/lustre/mhsieh/asym_memb/asym_memb_builder_v4_prot
or download the package through the link asym_memb_builder_v4.tgz asym_memb_builder_v5.beta.tgz
Authors: Min-Kang Hsieh and Jeffery B. Klauda
The demonstration for forming asymmetric bilayer from two dissimilarly sized monolayers, where the difference of surface area (X-Y plane) is more than 5%, is described as follows and corresponding files could also be found in the package. To read the lateral molecular structures of lipid, water, and ions, the input symmetric membrane system should be recentered by putting the lipid bilayer structure at the z = 0. The package provides a recenter procedure in the recenter folder. The asym_memb_bulider provides a semiautomatic executing script which could run the building process using CHARMM in two portions. By the commend ./run_builder_1.csh, the first portion includes Step 1.0 to 1.2 and generate an option table. Then, users could edit the step2_opt_a.str/ step2_opt_b.str to run the second portion by using the commend ./run_builder_2.csh, which includes Step 1.3 to Step 5.
Individual steps for building an asymmetric bilayer for NAMD simulation have been described in detail as follows in Instructions of each step.
Instruction of each step
Recenter procedure
@ ./recenter/
Edit 0_setting.csh to include toppar, psf, crd, and xst information of the bilayer which is desired to recenter.
Example of 0_setting.csh
top="/lustre/mhsieh/eng_memb/mol/crooks_p_run1/toppar.str" topdir="/lustre/mhsieh/eng_memb/mol/crooks_p_run1//toppar" psf="/lustre/mhsieh/eng_memb/mol/crooks_p_run1/step5_assembly.psf" crd="/lustre/mhsieh/eng_memb/mol/crooks_p_run1/step5_assembly.crd" xst="/lustre/mhsieh/eng_memb/mol/crooks_p_run1/namd/step7_production-16.xst"
By using the command ./0_setting.csh, symbolic links of toppar.str, psf and crd files as well as toppar folder are created, the unit cell parameters of the last step in the xst file are read to cryst.str, and the psfcrd.str is also generated.
Change the number of data points (frames) of the dcd file in recenter1.inp, recenter2.inp, and recenter3.inp.
set ts = 2500 ! number of data points in each DYN files
Include the dcd file (P) and the result name prefix (fname) in the 1_recenter.csh, 2_recenter.csh, and 3_recenter.csh
P=/lustre/mhsieh/eng_memb/mol/crooks_p_run1/namd/step7_production-16.dcd fname=crooks_p_run1
Then, using the command ./1_recenter.csh, ./2_recenter.csh, and ./3_recenter.csh sequentially.
Please find the detail in the part I of EDP analysis at https://user.eng.umd.edu/~jbklauda/wiki/doku.php?id=edp
Required files to begin
@ asym_memb_builder/ The recentered dcd, psf, xst, crystal_image.str and step3_size.str of input bilayers are required in the main folder. User should edit 0_setting.scr by providing the location of files. The 0_setting.scr includes these files by symbolic links using the follow command: ./0_setting.scr
Example of 0_setting.scr:
ln -s ./recenter/crooks_p_run1-recenter.dcd crooks_p_s1.dcd ln -s ./recenter/m2-o-run1-recenter.dcd m2-o-s1.dcd ln -s /lustre/mhsieh/eng_memb/mol/crooks_p_run1/step5_assembly.psf crooks_p_s1.psf ln -s /lustre/mhsieh/eng_memb/mol/crooks_p_run1/crystal_image.str crooks_p_s1-crystal_image.str ln -s /lustre/mhsieh/eng_memb/mol/crooks_p_run1/step3_size.str crooks_p_s1-size.str ln -s ../m2-o-run1/step5_assembly.psf m2-o-s1.psf ln -s ../m2-o-run1/crystal_image.str m2-o-s1-crystal_image.str ln -s ../m2-o-run1/step3_size.str m2-o-s1-size.str ln -s /lustre/mhsieh/eng_memb/mol/crooks_p_run1/namd/step7_production-16.xst crooks_p_s1.xst ln -s ../m2-o-run1/namd/step7_production-16.xst m2-o-s1.xst
Also, copy your toppor folder and toppor.str to here.
Step 1: generate monolayers
Step 1.0: check the input variables (co-executing with step 1.1)
Executing script: step1.0_input.scr
Output files:
1.step1.0_input_check.txt
2.step1.0_input_check.str: Parameter tables for later steps
Edit the step1.0_input.scr to include the information of the input symmetric membranes.
Example of step1.0_input.scr
f = "m2-o-s1" File name of Membrane 1 (M1) l = 78 Number of lipids per leaflet of M1 p = 2500 The frame selected from DCD file of M1 q = 10 frequency of DCD/frequency of XST r = "crooks_p_s1" File name of Membrane 2 (M2) s = 100 Number of lipids per leaflet of M2 t = 2500 The frame selected from DCD file of M2 u = 10 frequency of DCD/frequency of XST c = 0.15 conc. of KCL (M)
The default assumes that M1 and M2 would be the source of top and bottom leaflets, respectively, to generate the asymmetric bilayers. The parameter check could be performed individually through the command ./step1.0_input.scr, which calls step1.0_input.csh to read the parameters and automatically check the ratio of sizes of two input membranes (from the XST file by the given frame of trajectory). The input variables are copied to step1.0_input_check.str as a parameter table for later steps. Read step1.0_input_check.txt for the guide for the following steps. This step has been set to co-execute in Step 1.1 (called by system “./step1.0_input.scr” in step1.1_iso_monolayer.inp configuration file).
Step 1.1: Isolate monolayers
CHARMM configuration file: step1.1_iso_monolayer.inp
Output files:
1.step1.1_lipid_info.txt: the composition info of input monolayers
2.*.crd/psf/pdb files: the structural files for lipid/water/ion of input monolayers
In step 1.1, the operation of separating imported bilayer to two monolayers is as follows. First, the coordinates of top (0) and bottom (1) leaflets transfer their center of mass (COM) to the z-direction. Then, the bottom leaflet flips 180o along with the x-direction; that is, the head of lipids is toward the z positive direction and the tails are toward the z negative direction. Both monolayers align above the X-Y plan (z > 0). Two monolayers are saved as individual coordinate files.
Step 1.2: Provide new monolayer options with lipid compositions
CHARMM configuration file: step1.2_adding_lipid_options.inp
Output files: step1.2_adding_lipid_info.txt: the composition info of enlarged monolayers for selection.
In step 1.2, the small monolayer would be enlarged with a random centering point to equal the size of the other leaflet by adding lipids. First, the program checks the size of two imported monolayers. If more than a 5 % difference in the size (X length), the program executes the adding lipids process to the small monolayer. The operation of adding lipids is that (1) randomly selects the center point of the small monolayer and clones it to a n x n repeating cell in the x-y plane; (2) the surface is cut in the desired size (equals to the x length of the large monolayer); (3) the lipids outside the cutting area (v4: the phosphate of lipids; v5: the COM of lipids as referring for each lipid) are deleted to form a new monolayer. In the current version, users could decide how many monolayers they would like to generate for selection by changing the parameter Bulidmax in max_options.str (default is 5 for both top and bottom leaflets of the small bilayer). A list of options for new generated monolayer is stored in step1.2_adding_lipid_info.txt, which provide the composition of the new monolayer and the score of selection as follow,
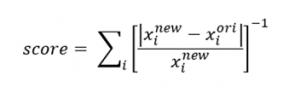
where xi is the mole fraction of the lipid i in the new and original monolayer. The score is the reverse of a sum of absolute relative deviation of the fraction, that is, the higher of the value, the higher similarity in the new and original monolayer in the lipid composition.
Example of step1.2_adding_lipid_info.txt
label option score xshift yshift #lip chg #type oype pmpe pmpg pope pype pypg qmpe M2-O-S1_0 1 1.4839 34.9284 42.5423 91 -11 7 5 38 3 6 22 8 9 label option score xshift yshift #lip chg #type oype pmpe pmpg pope pype pypg qmpe M2-O-S1_0 2 1.4043 26.9165 29.0049 100 -16 7 6 38 4 8 20 12 12 label option score xshift yshift #lip chg #type oype pmpe pmpg pope pype pypg qmpe M2-O-S1_0 3 1.4522 26.2889 17.7565 97 -13 7 7 40 5 8 20 8 9 label option score xshift yshift #lip chg #type oype pmpe pmpg pope pype pypg qmpe M2-O-S1_0 4 1.2647 42.7155 47.9648 99 -13 7 7 38 4 6 26 9 9 label option score xshift yshift #lip chg #type oype pmpe pmpg pope pype pypg qmpe M2-O-S1_0 5 1.1026 49.0564 42.4307 95 -11 7 6 39 3 6 25 8 8 label option score xshift yshift #lip chg #type oype pmpe pmpg pope pype pypg qmpe M2-O-S1_1 1 1.8604 46.4041 20.4853 99 -12 7 6 43 3 8 20 9 10 label option score xshift yshift #lip chg #type oype pmpe pmpg pope pype pypg qmpe M2-O-S1_1 2 1.1839 32.3908 30.0267 97 -13 7 5 41 4 6 20 9 12 label option score xshift yshift #lip chg #type oype pmpe pmpg pope pype pypg qmpe M2-O-S1_1 3 1.1371 32.2219 43.0549 97 -14 7 6 38 6 6 23 8 10 label option score xshift yshift #lip chg #type oype pmpe pmpg pope pype pypg qmpe M2-O-S1_1 4 2.3543 37.8645 33.7118 96 -14 7 6 37 4 7 22 10 10 label option score xshift yshift #lip chg #type oype pmpe pmpg pope pype pypg qmpe M2-O-S1_1 5 2.309 36.928 48.9222 99 -14 7 6 39 5 8 22 9 10
After selection from step1.2_adding_lipid_info.txt, the information (label, option, xshift and yshift) of selecting monolayer has to be stored step1.2_opt.str as an input to generate the molecular structure files.
Example of step1.2_opt.str
SET MEMBR M2-O-S1_1 SET OPT 4 SET Xshift 37.8645 SET Yshift 33.7118
Step 1.3: Generate new monolayer form the selected option
CHARMM configuration file: step1.3_adding_lipid.inp
Output files:
1. step1.3_adding_lipid_check.txt: double check the composition info of selecting monolayer
2. step1.3_adding_lipid_m2-o-s1.txt: the operation summary including the original and new generated monolayer structural info.
3. step1.3_adding_lipid_option.str: the input parameters for the next steps.
4. Adding_lipid_*.crd/psf/pdb files: the structural files for lipid/water/ion of new generated monolayer.
Step 2: Generate a bilayer
CHARMM configuration file: step2a_gen_bilayer.inp (with adding lipid) or step2b_gen_bilayer.inp (without adding lipid)
Output files:
1. Step2_bilayer_*.crd/psf/pdb files: the structural files for lipid/water/ion of new generated bilayer.
2. step2_gen_bilayer.str: Parameter table of the generated bilayer
In step 2, the two monolayers listing in the input file (step2_opt_a.str or step2_opt_b.str) are combined to form a bilayer. If adding lipids, step2a_gen_bilayer.inp is used with step2_opt_a.str as the input; otherwise, step2b_gen_bilayer.inp is used with step2_opt_b.str as the input. The centers of mass (COM) of both monolayers line on the z-direction.
Step 3: Determine size of simulation box
CHARMM configuration file: step3_size.inp
Output files: step3_size.str: Parameter table of the simulation box
In step3, the size (z length) of the simulation box is determined to refer to the amount of water which includes in the system. For the size of the simulation box, the larger X (=Y) length would be used to set the system of asymmetric bilayers. Note that the total charge and the ratio of water to lipid are shown in the step3_size.str. The ions (default: KCl) concentration could be changed by editing the CONC value (default: 0; only neutralizing ions)
Step 4: Form molecular structures of each part in simulation box
Step 4.1: Generate lipid structure
CHARMM input file: step4.1_lipid.inp
Output files:
1. step4_lipid*.crd/psf/pdb file: the structure lipid/water/ion of bilayer simulation box
2. step4_components.str: water and ions count info
In step 4.1, the input step2_bilayer*.psf/crd files (internal setting) would be imported and examined the structure by separating lipid structures from water and ions if they exist. It also could be used for processing the structures with other molecules (e.g., proteins) in this module.
Step 4.2: Generate water structure (inactive; default: directly use the input lateral water)
Step 4.3: Generate ion structure
CHARMM configuration file: step4.3_ion.inp
Output files:
1. step4.3_addnneg.crd and step4.3_addnpos.crd: the adding ions coordinate files.
2. step4.3_ion.pdb: the final ions structure file.
3. step4.3_ion.str: Parameter tables for ions.
In step 4.3, if the neutralizing ions only (CONC = 0), the number of ions will be adjusted by using a faster distance-based algorithm to add more ions or random pickup to remove the extra existing ions. If KCl conc. is non-zero, the extra ions added to the system for the setting concentration after addition the neutralizing potassium ions.
Step 5: Assemble and run
step5.1: assemble
CHARMM configuration file: step5.1_assembly.inp
Output files:
1. step5_assemble* file: the assembly bilayer structure
2. step5_assembly.str: the final info of simulation box.
In step 5, the bilayer, water box, and ions are assembled into the simulation box.
Step5.2: Generate NAMD input
Update:
One should be able to create input for several MD simulation packages by Force Field Converter at CHARMM-GUI page (https://charmm-gui.org/?doc=input/converter.ffconverter)
where takes the step5_assembly.psf and step5_assembly.crd file generated from step5.1 with step5_assembly.str information.
CHARMM configuration file: step5.2_input.inp
Output files: ./namd directory
In this step, the assembly structure is prepared for running step 6.1 to 6.6 equilibrium using NAMD. The package adapts the previous approach from CHARMM-GUI that the system is pre-minimized using CHARMM with the membrane restraint files. All required files are included in the package as follows:
1. step5_input_minimization.str 2. crystal_image.str 3. membrane_restraint.str 4. membrane_restraint2.str 5. namd/step5_charmm2namd.inp 6. namd/membrane_restraint.namd.str 7. namd/membrane_restraint2.namd.str 8. namd/step5_charmm2namd.colvar.str
The required python scripts for processing are also included in the package as follows:
1. checkfft.py 2. convert_par2namd.py
After completion of step 5.2, a simulation package (./namd/) for NAMD is prepared with a short energy minimization of membrane system under restraint (see step5_input_minimization.str and membrane_resitraint.str files) and ready for the step6.
Step 6: Equilibrium run
In this step, the charmm2namd pdb/psf files and NAMD configuration files are prepared in ./namd/ folder for running step6.1 to step 6.6 equilibrium.
Step 7. Production run
A production run template is provided.
