Table of Contents
Brief Instruction to calculate electron density profile of multi-component lipids
Author: Xiaohong Zhuang Updated (2021): Dr. Jeffery Klauda Updated (2021): Robert Allsopp (Combining the .sim and renaming atoms) Updated (2023): Abhi Senthilkumar (Python scripts to automatically rename atoms and combine .sim files into xlsx) Updated (2023): Joshua Lucker (Clarification on Step 6)
Example of analysis: e.g._thickness.xlsx
The scripts that you need are in the following paths:
ZT1 path: /afs/shell.umd.edu/project/energybio/shared/jbklauda/scripts/edp_mult_lipids or using the zip file: edp_mult_lipids.tar.gz
Also if there is a protein download the protein_analysis.tar files and run them on as many proteins as there are in the system, and combine the data similarly to before
Part I. (Recenter bilayer)
This section is for recentering the lipid bilayer so that it is always at the center of the box. This is essential so that the density profiles have the same reference (minimum bilayer density is a z=0).
Copy the directory /afs/shell.umd.edu/project/energybio/shared/jbklauda/scripts/edp_mult_lipids into your namd directory with the DCD files.
1. Edit cryst.str file based on your crystal data (A,B,C,Alpha,Beta,Gamma) in dyn.xsc which is the system size of last frame in dyn directory. box.cry file is same for all lipids, so no need to edit.
2. Edit recenter*.inp files.
What you need to change is the following in the recenter1.inp:
set fdyn = 11 ! First traj. in series set ldyn = 12 ! Last traj. in series set ts = 500 ! Number of data points in the DYN files ( ts = numsteps/dcdfreq) (more lines ...) open read unit @ru file name ... ! Read original trajectory (update!)
fdyn and ldyn are the first and last dyn of your equilibrated system. Same file range (first and last traj #) will need to be changed in the recenter2.inp and recenter3.inp files as well as the ts value. ts is obtained by reading your dyn-2.inp file to find dcd frequency and total time steps=numsteps/dcdfreq. Also, don't forget to update the trajectory template to you want to read (only in recenter1.inp), edit the following line:
open read unit @ru file name ../../dyn@j.dcd ! Read original trajectory
3. Update the path of files in the psfcrd.str to what you used for the simulation
4. Copy the toppar.str and toppar folder into the recenter directory. This should be from the root charmm-gui folder where the CHARMM input files are located.
5. The recenter*.csh files call the CHARMM executable using the most up-to-date code. If using off DT2, this will need to be changed.
Once you've made the changes, submit the three recenter#.scr in order and only after the previous has finished using commands: recenter1.scr, and then run recenter2.scr, recenter3.scr
Make sure one file is finished running before doing another one. This will create new DCD files but the only one you will keep are the dyn#-recenter.dcd because once your run *3.csh the *tmp.dcd files will be deleted.
Part II (Density Calc)
The next step is to take these re-centered DCDs and calculate the densities for each atom in the bilayer. Read the topology file to generate the *atmlbl.txt and *cmp, in order to specify the groups of the atoms in your system.
1.1 Update the lipid name in 1_gen_rtf_atmlbl_cmp.scr:
foreach lip ( DGPE )
1.2 Copy the toppar.str and toppar subdirectory in the recenter directory.
1.3 The list contains all the lipid topology files currently exist. You may need to add the topology file name if your lipid is in a new topology file that you generated. Add it to the list in the foreach section shown below of the 1_gen_rtf_atmlbl_cmp.scr file
foreach rtf (${p}${j}lipid.rtf …
${p}1_toppar_cua_lipid_7_12_15.str'' **Add_new_topology_file_here)**
1.4 Run the script by command 1_gen_rtf_atmlbl_cmp.scr.
NOTE: Difference in atmlbl.txt and cmp between 1-lipid and multiple-lipids:
| 1-lipid system | Multiple-lipids system |
|---|---|
| 2nd column of atmlbl.txt is segid MEMB | 2nd column of atmlbl.txt is resname |
| Water atoms are the last 3 atoms in atmlbl.txt | Water atoms are in a separated atmlbl.txt |
| Water(Wat) is the last group in cmp | Water(Wat) is in a separated cmp |
2. Get atom data
2.1 Edit the getatomnd*.inp files and 1-run*.csh files (specifying the range, changing the charmm program used in the .csh file as described in Part I, Step 5, etc)
set ff = 11 ! file range: first dcd file set lf = 12 ! Last dcd file to use set zin = 0.2 ! slab thickness (no need to change) set mxz = 50 ! max z value, should be slightly (maybe 3~5) greater than the C/2 (3rd number/2 in cryst.str or dyn.xsc). e.g. if C is about 90, then mxz 50 (=90/2+5) is used. … calc mxf = 500 * @N ! Update number of frames in each dyn file (=#total time steps divided by dcd frequency)
2.2 Update the psfcrd.str file to have the correct location of the psf and crd file.
2.3 Update lipid (file) name in components.scr
Note: components2.scr and 1_run2.csh are not needed for systems with only a few kinds of lipids. Also, use lower case for the lipid names not upper case.
2.4 Run the scripts by ./1_run.scr which will calculate the EDP of the lipid.
The output *.dat amd *out.gz files found in the atoms subdirectory (Don’t worry if you don’t have it now, the 1_run.scr will make it for you).
Part III (Master Density)
Combine atomic density data
And then combine the density *dat files to obtain combined EDP that can be used by a program SIMtoEXP (see detail later) that gets you the density profiles.
Run the simulation to combine the ~.dat files to obtain popc.sim file. *.x file may not be executable for the first job, so first type the following to make it executable:
chmod u+x bldsim.x
then run the script buy command: ./bldsim.x *-atmlbl.txt ./atoms_* *.sim
(replace * by your lipid name). e.g. ./bldsim.x dype-atmlbl.txt ./atoms_dype dype.sim
Please make sure include “.” and space due to the way that bld-sim.f is written). This will create *.sim files of all lipid components that has the combined atomic density which will be used in SIMtoEXP.
New method of renaming the atoms and compiling in an excel file
- First edit the script format.py. Edit the LIPID_NAMES and LIPID_ABBRS lists. The script automatically changes the .cmp and.sim files, but this can be changed by editing the FILE_ENDINGS list. One side effect is that the comments at the start of the file will also be targeted by the regex, but this effect is minimal. The script will substitute all 'z's in atom names with a 26. If there is a different substitution you would prefer you can change that. You can also choose to either modify the original .sim files or create new formatted files. It is recommended to leave MODIFY_FILES to False to avoid having to regenerate the .sim and .cmp files if something goes wrong. Once you have edited the script, make sure you have loaded python with module load python. Then run the script with python format.py
- Next, edit the script generate_excel_file.py. Edit the LIPIDS list to add the lipid.sim files you want to compile. If in the previous step you generated new files, make sure to set USE_NEW_FILES to True. If this is the first time you have run this, you will need to install xlsxwriter for python. Do this with the following command: pip install --user XlsxWriter. Then, run the script with python generate_excel_file.py
- This will generate an excel file titled sim_data.xlsx along with .csv files for each lipid.
Old method of renaming atoms and compiling in an excel file
*Then combine the individual .sim files in excel into one master file that contains all of the individual lipids, peptide, water, ions. To do this copy everything from the first file including the Z column and then from that file onward exclude the Z column.
*Be sure to rename all of the atoms that have the letter z anywhere in the atom name as the simtoexp ignores any atom column with a z anywhere, also be sure to rename every single duplicate atom name in the .sim file. No overlapping atoms can occur. To prevent overlap it is important to first start by adding an extra letter specific to the lipid type onto the end of every atom so POPE e, POPG g, TOCL2 T… Then check through by copying the row of atom names and transposing them then use in excel filter sort A-Z, and then use the code =if(QQ5=QQ6,1,0), to check through and flag down any duplicate names. Then be careful to rename them also in the .cmp file.
*Third for atoms like potassium or chlorine ions or other non-standard atoms it is a good idea to check that the it isn't being confused in simtoexp. Simtoexp uses the first letter of each atom name to identify what type of atom it is c-carbon p-phosphorous h-hydrogen and the rest of the atom name is used to differentiate the different atom names. To manually adjust the names go to tools→scattering lengths and add k for potassium as an example at the bottom instead of one of the numbers, enter the number of electrons. Then go to tools→atomic form factors switch to the newly created k tab and input the values of the potassium ion from the link below.
http://lampx.tugraz.at/~hadley/ss1/crystaldiffraction/atomicformfactors/formfactors.php
Part IV. (Verify)
It is useful to verify the calculated EDP because a minor mistake can be easily missed and then the edp may be totally off.
There are two types of scripts to verify the No. of atoms: 1. single-type lipid 2. Multi-types lipids
To run the scripts, go to verify_Ne_mult_lipids directory, and make the following updates:
1. Update the lipid name in run_check.scr (treat ions (POT and others) and water (tip3) as lipid components)
foreach lip ( l2pc plpe pot tip3 ) # Update the lipid name based on your sim and cmp file name
2. Update the following information in update_lipid.py :
N_lip = {'l2pc':22,'plpe':13,'pot':9,'tip3':1500} #lipid name in LOWER CASE and number of each lipid PER LEAFLET water/ions are total/2 (per leaflet)
SA_tot = **2079.0** # TOTAL SURFACE AREA (SA/lip * #lip/leaflet)
3. Submit the script by command: ./run_check.scr
The formula used to verify is N= Sum (density*Area* bin size 0.2), and N calculated from *sim should equals to the expected total atom/electron number.
NOTES:
a) Python script is case-sensitive, make sure the lipid names are same as in *.sim,*.cmp
b) Python has strict line indentation format, make sure not adding any additional space in the beginning of each line, but any space after 2nd character of each line is fine.
c) May need to update the path in run_check.scr if the sim and cmp are not in one directory out.
d) Make sure the group names in your *cmp agree with groups names in file E_data.py (which are also shown in Part II.2). If your group does not exist yet, please add your group names and number of atoms and electrons into E_data.py N_aeh dictionary.
e) In *cmp, the second column are the total No. of atoms of each group.
f) In *cmp, the non-comment contents start from 5th line. ( sed '1,4 d' )
Both types of scripts check the following in each part and the results is shown in check_*.txt:
1. Total No. of atoms in *cmp based on *sim
EXACT: the total number of atoms based on the numbers on the 2nd column of cmp
CMP: the total number of atoms based on the number of atom names in cmp
SIM: the total number of atoms based on the number of atom names in sim
2. Atom names mismatch between *cmp based on *sim The error message will be shown if an atom exist in one file but does not in the other. Part 4 will automatically stop due to atom name mismatch error.If this occur, please fix the atom name(s) in *cmp or *sim.
3. Zero density of an atom This checks if any atom has the zero total atomic density in *sim. A warning will show if an atom (including the atom name) has the zero atomic density.
4. No. of atoms and electrons in each group *sim In each of atoms and electron part, four columns will be shown: Group names, Expect (Number of atoms per group (Head group based on internal library data, the tail groups based on the numbers on the 2nd column of cmp), CMP (the total number of atoms per group based on the number of atom names in cmp),SIM (the total number of atoms per group based on atomic density in sim). A warning will show if the Deviation fraction of electron No. exceeds 0.01!
=======================================================================================
If all you need is to calculate bilayer thicknesses, please see the note in Part VII. Part V and VI contain lots of extra information, and the use of SIMtoEXP is not always necessary.
========================================================================================
Part V. (SIMtoEXP)
Obtain SIMtoEXP software in using zip file: simtoexp.zip. It does not work in linux system. It has user graphical interface and works on Windows only (Unfortunately, there is no Mac version).
Read the manual for the SIMtoEXP code and then
1) Load the *.SIM and *.CMP file to see density plot in SIMtoEXP. Update/Enter the ED_wat and NSLD_wat based on the values of water level off in Electron Densities and Neutron SL Densities windows, respectively, click the fix’s, and then click Fourier Transform and Calculate Volumes each time loading the sim and cmp files.
SIMtoEXP can also easily check that whether the atoms in *.sim and *.cmp match: load *.sim first, and then load *.cmp file, and finally load the *sim again, the SIMtoEXP will show the atoms that mismatch in these two files if there is any. This step is probably not necessary. The atom mismatch is checked in Part III.
2) After you load *.SIM and *.CMP file into SIMtoEXP, in the ED plot, find the z values (positive and negative) values that beyond which that water ED suddenly drop in ED plot which meaning it is beyond the membrane, or level off region of ED. Then manually remove the data in *sim file. For POPG, it is regions z < (about -28 Å) and the region z >( about 28 Å). 3) Then use the new cut *sim file for comparison to experimental XFF and NFF data.
4) Export the data and then plot it in Excel, Matlab or other software.
After the EDP is obtained, the data has both negative and positive z (usually -36 to 36 as we set the max z range). However, edp is usually shown with positive z only. The total electron density data/plot can be symmetrized(averaged) by using (EDP Value on positive z + Value on negative z)/2 and then plot the average data with the positive z only. (This is be done by export the edp data and imported into Excel. Then sort and then copy the column of negative values to the column next the positive values. And then calculate and plot the average).
Part VI. (Compare)
The files needed are popc.sim, popc.cmp ,(the *.sim and *.cmp files of your lipid), and also the experimental Xray and Neutron FF data of your lipid. Do not do this section if you lack experimental data to compare.
1) In SIMtoEXP, go to file → OpenSIM and OpenCMP to open the popc.sim and popc.cmp files respectively. Then click file → OpenEXPx → SAM1 to open the experimental x-ray scattering file. and then OpenEXPn→SAM1, SAM2,SAM3 respectivey to open the the experimental neutron scattering file with 3 contrast(100%D,70%D,50%D).
Note (please read): when the each neutron scattering file is opened, the scattering length window will pop out with automatically updated NSL (neutron scattering length) of D which are defined in the experimental data file. If the window doesn't pop out, go to Tools → Scattering Length. For example, for 100%D, D=6.67e-5 (this depends on the water density). If standard 1 g/cm3 is used, then D=6.38e-6 Angs^2; 50D means 50%D2O and 50%H2O, (D=frac_water*D(water) + frac_d2o*D(D2O)). For water, note that D(water)=-5.58e-7 at 1 g/cm3. Therefore, when a different deuterium contrast (??D) file is opened, NSL are usually automatically updated based on the definition of D. To verify this, open a 50D NFF experimental data, it shows something like:
## redefinition of scattering power for D such that water corresponds to 50% D2O
#SLwin
#set b(1) 1.465e-005
#setRHO_wat N 2.915e-006
As a note if this does not occur, go to the scattering length window and change the deuterium number in the NSL column manually based on the equation: D=frac_water*D(water) + frac_d2o*D(D2O), where D(D2O)=6.67e-5, and D(pure water)=-3.74e-5. Then, change the NSLD_wat number in the main window based on the maximum value in the Neutron SL Densities window (Note: you can also use the above equation where D(D2O)=6.38e-6 and D(water)=-5.58e-7 as well if you wish).
2) Update/Enter the ED_wat and NSLD_wat based on the values of water level off in Electron Densities and Neutron SL Densities windows, respectively, click the fix’s, and then click Click Fourier Transform, and Calculate Volume. (Make sure to click these two icons each time a new file is opened/loaded.) Go to X-ray FFs and Neutron FFs to compare the data. I recommend you make sure your plot is reasonable before you export the data and plot. POPG may behave like POPC, so the curves will be probably similar.
Part VI and 1/2: Create own comparison plots
We want to re-plot since the graph in SIMtoEXP is not well formatted for publishing.(e.g. Only one curve is showing at one time; the axe titles are missing etc.).
1) Export the simulated X-ray FFs and Neutron FFs data from SIMtoEXP by go to Tools→EXPORT→ ALL. Only the D% data shown with the curve will be exported. To export each D constrast, We need to select each by click on the space in SAM# and scale, and hit enter key. The simulated data of that D% will be plotted and exported. Each export one D%. If there are three D%, it would require three export.
2) You can plot in Excel or Matlab. If you use Matlab, copy and paste the exported simulated data to Matlab data file. And define the columns correctly. The experimental xray and neutron data are rescaled to reach the minimum Chi Square (fitting with simulated data) as shown in SIMtoEXP. (The re-scale does not affect the comparison of the FF data). When you plot, you must multiply the experimental data with the scale shown in SIMtoEXP for both Xray and Neutron data. And then plot the simulated data and re-scale experimental data in Matlab. I multiply the scale in Excel and then use the calculated rescaled data in Matab. But you can just simply multiply the scale in Matlab and define it as y values. When plot form factors comparison graphs, plot experimental data first, use hold on, and then plot simulated data, so that simulated curve can superimpose on the experimental data, which is in better view.
The experimental data used in SIMtoEXP is adjust the % of D2O by automatically fix the NSL of D (as you can see them after load the different D2O% samples and go to Tools–>Scattering Lengths)
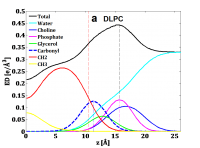
The MD-based electron density profiles of DLPC at 50 °C. This is averaged electron density profiles obtained by (EDP Value on positive z + Value on negative z)/2. The original EDP that you have EDP in both both the positive and negative z. (Please ignore the vertical dash lines. The original electron density profiles does not have them. )
Part VII. (Thicknesses)
========================================================================================
Note: This can be done more easily using the directory “thickness_mult_lip”.
1. 1_lipid_edp.scr needs to be updated for lipid type (you may not want to include 'tip3' and 'pot' if you are going to run 2_comb.scr since the later will add 'tip3' and 'pot' for you). Then run *.scr files in order.
2. The gnu file may need to be updated for column selection. This is only for plotting and may not be necessary.
3. thickness.py may also need to be updated if the interval contains extraneous maxima. Adjust the interval by changing “Nn”.
========================================================================================
Thickness estimate with SIMtoEXP
Another way to obtain thicknesses is from the SIMtoEXP program (separate from the python code listed above).
1. Load *sim and *cmp files into SIMtoEXP by: GUI–>File–>OpenSIM–>*.sim , and similar for *cmp.
2. Read the values of the electron density of water and the neutron scattering length density of water and enter these values on the GUI in the ED_wat and NSLD_wat boxes, respectively. (This step is important for XFF and NFF. It may not affect volume probability, but I suggest to do it just in case.)
3. Then three thicknesses can be obtained by reading the values in graphs:
3.1 The hydrophobic thickness (DC) is estimated by the distance between the midpoints of bilayer's hydrocarbon acyl chains based on volume probabilities;
Therefore, in the Volume Probabilities graph, go to ToolAn→alysis;
Combine all three types of carbon group together by enter CH,CH2,CH3 and then press Combine Components, then the combined curve will show in the graph.
Find the point of VP=0.5 by zoom in (Manual point 14 to 17 are shown below in green). The very exact 0.5 may not be necessary, but try to get as close as possible. I usually zoom in 3 or 4 times to get x-coordinate value which is the Z in the bilayer. Read both the positive and negative values. 2*DC=DR-DL.
The following is part of the SIMtoEXP manual that is provides helpful tips:
“ 14)Manipulate the axis: GUI–>Axis
‘show more x/y’ expands x/y axis by 10% on each end,
‘show all’ sets the axis automatically to show all the data points.
15)Locate the crosshairs on the GUI and left click. The top right of the GUI gives x and y values.
16)You can zoom by locating the crosshairs, hold down the Ctrl key, drag the mouse to outline a dashed zoom box and then left click. To unzoom, Ctrl right click.
17)Left clicking on the graph elements in the legend to the right of the GUI toggles them. Right clicking reorders which curve appears ‘on top’ in the display. Ctrl+Left click removes the curve from the graph.” (SIMtoEXP manual)
The similar method is applied for DB and DHH. The only difference is there is no need to combine components.
3.2 The overall bilayer thickness (DB) is found from the distance between the midpoints of water based on volume probabilities;
3.3 The headgroup-to-headgroup distance (DHH) is the distance between the peaks of total electron density in the EDP graph. (Here is the peaks distance, not the midpoints distance)
Please refer to Excel file eg_thicknesses.xlsx for the example measurement.