Protein Contact Map
(author: Dr. Min-Kang Hsieh)
This brief instruction is used to generate protein contact maps by a VMD script. The VMD script is run in Deepthought instead of VMD software.
The scripts are in the DT2 path : /lustre/mhsieh/contactmap/
or using the zip file:contactmap.gz
1. Please copy the script folder(s) for your protein(s) from the path highlighted above to your directory that has all the dyn files.
2.1. In 1_contact.tcl, update the parameters (d is the cutoff distance, i is the first trajectory file and f is the last trajectory file) for your calculation.
set d 50
set i 6
set f 10
2.2. Update the path for your psf and trajectory (xtc or dcd) files (If they are in a different path). Note that k will go over all trajectory from the first to the last files.
set psf ../step5_assembly.psf
set xtcFile($k) ../gromacs/step7.${k}_production.xtc;
2.3. Run ./1_contact.scr to submit your job for vmd calculation (calls 1_contact.tcl and then calls newcontacts.tcl) and calculate residue pair distances.
3.1. After the vmd job is completed, update the parameters in 2_run_map.py, where num_res is the number of residues, num_frame is the number of total frames from trajectory files, max_dist is the maximum distance which used in demonstrating the maps.
num_res=43
num_rows=num_res
num_columns=num_res
num_frame=20006
max_dist=10
3.2. Run ./2_run_map.scr to generates data files (map_frequency.dat and map_distance.dat) for drawing maps.
4. Run ./3_run_dist.gplot to generate two protein contact maps which show the frequency and the average distance of contact, respectively.
Note
Removal non-physical spots on the map
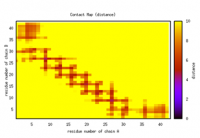
Here is an example (see the figures below) for comparison of maps before and after screen off non-physical spots. Frequency calculation collects frequency counts for distance < 10 Å.

The average distance for individual residue pairs (cutoff distance is 50 Å, which includes almost pairs in a whole trajectory) shows some spots in the upper right concern with low percentages. This is not physical and may be the result of only a few frames causing this.
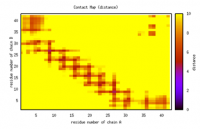
Removing low percentage levels (e.g., below 25%) of contact in the distance map, which only considers the percentage levels as contacts within the distance below 10 Å. The default setting in the code is to only consider distances that occur > 25% and the upper limit in the distance map is 10 Å. The result show as follows.